Abstract
Introduction:
Somatic mutations are common in MDS and mutation profiling is becoming increasingly standard as part of the initial diagnostic evaluation. While clinical reports often indicate the predicted prognostic significance of specific mutations identified, they do not predict the degree of clinical impact or how these predictions actually correlate with clinical outcomes. We investigated the utility of the Ensembl Variant Effector Predictor (VEP) tool in correlating specific mutations with outcomes in MDS.
Methods:
MDS patients from the MDS CRC with available molecular genetic data by next-generation sequencing were included in the analysis. Mutations present at diagnosis were obtained, converted to HGVS notation and then categorized using the VEP tool into mutations with predicted high, moderate, or low impact based on sequence variants and structural variants including copy number changes or insertion or deletions. (McLaren , Genome Biol. 2016). We also utilized SIFT, which predicts amino acid substitution affect on protein function based on degree of conservation of residues. Neither tool accounts for VAF. The mutation categories were correlated with survival in a univariate log-rank test. Complete remission (CR) status was defined per IWG 2006. Cox proportional hazards model of time from diagnosis to death or loss of follow up with mutation category, age, IPSS classification was applied.
Results:
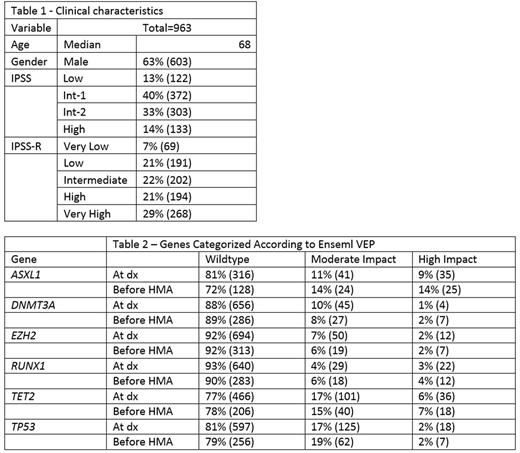
Clinical characteristics of the 963 included patients are summarized in Table 1. We categorized mutations of the following 6 prognostic genes, identified within 6 months of initial MDS diagnosis, using Ensembl VEP: ASXL1, DNMT3A, EZH2, RUNX1, TET2, and TP53. The distribution and predicted impact of the mutations are summarized in Table 2. None of the mutations were predicted to have a low impact. In our cohort, onlymutations in DNMT3A and TP53 were correlated with worse overall survival (OS). (HR 1.67, p<0.001, CI=1.26-2.22, and HR 2.46, p<0.001, CI=1.97-3.08, respectively). For DNMT3A mutations, worse OS was associated with mutations categorized as high impact (HR 2.35, p=0.012, CI 1.2-4.59), but not with those termed moderate impact (HR 1.33, p=0.159, CI=0.90-1.97). Among TP53 mutations , though, adverse OS was associated with both high impact (HR 2.63, p<0.001, CI=1.53-4. 54) and moderate impact mutations (HR 2.35, p<0.001, CI=1.78-3.11). High impact DNMT3A mutation was associated with worse OS (HR 2.41, p=0.01, CI=1.22-4.8) independent of age and IPSS cytogenetic risk, while moderate DNMT3A mutation was not associated with OS (HR 1.25, p=0.28, CI=0.83-1.87). SIFT predictions were not associated with OS in the genes individually, however presence of 2 or more genes predicted to be deleterious by SIFT was associated with poor OS adjusted for age, IPSS cytogenetic risk (HR 1.91, p<0.01, CI=1.17-3.12).
The mutational profile of the six genes was characterized in 270 patients tested within 6 months of starting treatment with azacitidine or decitabine. The distribution and predicted impact of the mutations are summarized in Table 2. Similar to mutations characterized at diagnosis, worse OS was associated with high impact DNMT3A mutations (HR 3.39, p=0.002, CI 1.57-7.34), but not with mutations predicted to have a moderate impact (HR 1.16, p=0.63, CI=0.63-2.17). Among patients with TP53 mutations treated with hypomethylating agents, adverse OS were associated with moderate impact mutations (HR 2.23, p=<0.001, CI 1.52-3.26), but not high impact mutations (HR 1.51, p=0.42, CI=0.55-4.11). Among patients who achieved complete remission, only moderate impact RUNX1 mutations (HR=0.16, p=0.03, CI=0.01-0.84) and high impact TET2 mutations (HR=0.17, p=0.025, CI=0.01-0.90) were negatively associated with CR. Moderate impact RUNX1 mutation and high impact TET2 mutation however were not associated with OS. No other mutations were associated with achieving CR after treatment with hypomethylating agents.
Conclusions:
While testing for myeloid-associated mutations is becoming standard practice as part of the diagnostic evaluation of MDS patients, interpretation of the sequencing results is complex and may be subject to technical variations that are obscure to clinicians. Presence of an abnormal mutation by itself is not enough to predict prognosis; predicted impact along with clinical characteristics need to be incorporated to establish their relative impact on clinical outcomes.
Lee: Clinipace: Consultancy; Baxalta: Consultancy; Alexion Pharmaceuticals: Consultancy; Amgen: Consultancy. Komrokji: Celgene: Honoraria; Novartis: Honoraria, Speakers Bureau. Steensma: Novartis: Consultancy, Membership on an entity's Board of Directors or advisory committees; Pfizer: Consultancy, Membership on an entity's Board of Directors or advisory committees; Celgene: Consultancy; Onconova: Consultancy; Janssen: Consultancy, Research Funding; Incyte: Equity Ownership; H3 Biosciences: Consultancy; Takeda: Consultancy; Amgen: Consultancy, Membership on an entity's Board of Directors or advisory committees; Pfizer: Consultancy. Ritchie: Pfizer: Consultancy, Other: Research funding to my institution; NS Pharma: Other: Research funding to my institution; Bristol-Myers Squibb: Other: Research funding to my institution; Astellas Pharma: Other: Research funding to my institution; Novartis: Consultancy, Other: Research funding to my institution, and travel, Speakers Bureau; Incyte: Consultancy, Speakers Bureau; Celgene: Consultancy, Other: Travel, Speakers Bureau. Hassane: Cellectis: Research Funding. Sekeres: Celgene: Membership on an entity's Board of Directors or advisory committees. Roboz: AbbVie, Agios, Amgen, Amphivena, Array Biopharma Inc., Astex, AstraZeneca, Celator, Celgene, Clovis Oncology, CTI BioPharma, Genoptix, Immune Pharmaceuticals, Janssen Pharmaceuticals, Juno, MedImmune, MEI Pharma, Novartis, Onconova, Pfizer, Roche Pharmace: Consultancy; Cellectis: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.